SARS-CoV-2 nanopore sequencing by NextGenPCR®
‘Midnight protocol’
Wouldn’t it be great to decrease PCR library time for SARS-CoV-2 nanopore sequencing from 3.5 hours to 21 minutes?
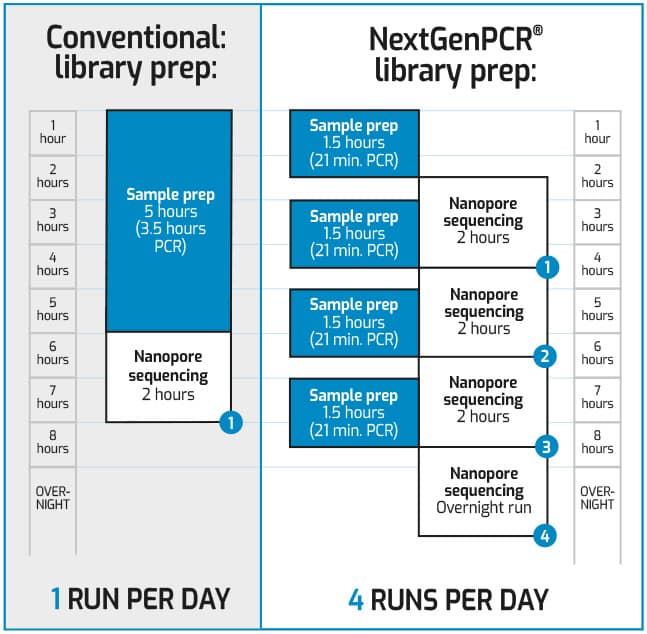 Genomic epidemiology is an important factor in the public health battle against COVID-19, enabling rapid identification and complete characterization of SARS-CoV-2. Combining the Midnight protocol developed by the Artic Network with NextGenPCR® technology gives you sequencing results for 96 samples in 3,5 hours instead of 7 hours. Thereby, using NextGenPCR® allows you to perform 4 runs per day instead of 1 run per day in the current workflow. It increases the throughput from 96 samples to 384 samples per day.
Genomic epidemiology is an important factor in the public health battle against COVID-19, enabling rapid identification and complete characterization of SARS-CoV-2. Combining the Midnight protocol developed by the Artic Network with NextGenPCR® technology gives you sequencing results for 96 samples in 3,5 hours instead of 7 hours. Thereby, using NextGenPCR® allows you to perform 4 runs per day instead of 1 run per day in the current workflow. It increases the throughput from 96 samples to 384 samples per day.